ImagesReslice
Path
[Images|Reslice]
Usage
The following submenus allow to reslice a volume in the transaxial, sagittal or coronal projection. This concerns not only the display of images but also includes the change in dimensions and other related parameters. The orthogonal reslicing used is based on the number of pixels or slices in each dimension, meaning that the original volume and number of voxels is preserved. To further protect the study integrity, a number of limitations or restrictions have been made:
- only reconstructed image data
- identical image sizes and pixel types required
- original volume should be at least ten slices thick
Remarks
- The viewer scales the images to the aspect ratio of the real world dimensions.
- Slice overlaps/gaps are masked using the centre-centre slice separation.
- A transaxial (XY slice) orientation is assumed in case of an unknown initial orientation.
- Because the number of projections can change after reslicing, we do addapt the time per projection parameter to hold an identical heart rate within Gated SPECT studies.
The following figures show how the slices are cut from the volume. The small axes represent the monitor screen dimensions (X=horizontal, Y=vertical). For each new slice orientation the origin and new screen X',Y' mappings are shown below.
| Transaxial | Sagittal | Coronal |
|---|---|---|
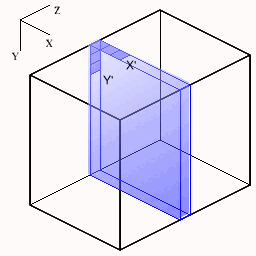 | 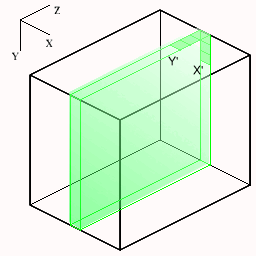 | 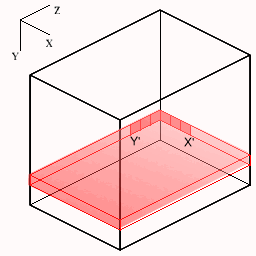 |
<< Extract | MenuWindow | Flip >>
